

Document Type : Original Article
Authors
1 Surgical Research Society (SRS), Students' Scientific Research Center, Tehran University of Medical Sciences, Tehran, Iran
2 Azad University of Medical Sciences, Tehran, Iran
3 Department of Medical Genetics, Afzalipour Faculty of Medicine, Kerman University of Medical Sciences, Kerman, Iran.
4 Division of Applied Bioinformatics, German Cancer Research Center DKFZ Heidelberg
5 Department of Computer Engineering, University of Kashan, Kashan, Iran
6 Urology Research Center, Tehran University of Medical Sciences, Tehran, Iran
Abstract
Graphical Abstract
Highlights
Keywords
Main Subjects
Introduction
Autosomal Dominant kidney disease is a condition affecting the entire body, caused by a mutation in PKD1 or PKD2 genes located on chromosome 16 or 4, respectively. These genes are responsible for encoding PC1 and PC2 proteins, known as polycystin-1 and polycystin-2 (1-3).
Malfunctions in the functions of PC1 or PC2 result in the disruption of growth regulation, G protein functions, and both canonical and non-canonical WNT pathways (4). The second hit model states that additional somatic mutations lead to increased cyst growth. In addition to the normal renal involvement with the progressive expansion of the cyst that leads to the enlargement and extensive distortion of the kidney structure and finally to the final stage of kidney disease, multiple extra-renal manifestations such as cysts in other organs, diverticulosis, abdominal and inguinal hernia, vascular malformations (5, 6). Renal cancer is a prevalent type of cancer globally, and its occurrence is on the rise. The primary histological types comprise clear cell RCC (ccRCC), chromophobe RCC, and papillary RCC (chRCC) (7, 8). A systematic review of 14299 primary renal tumor samples and 969 metastatic samples revealed a trend toward the increases of copy number alterations (CNAs) for both losses and gains and a significant difference between primary tumors and metastatic linked to BAP1 (9, 10). A majority of patients with RCC have an inactivated von Hippel-Lindau (VHL) tumor suppressor gene (11-14). Mutations involved in RCC lead to metabolic dysregulation, intramural heterogeneity, angiogenesis and harmful tumor microenvironment (TME) interference (15).
The Cancer Genome Atlas Research Network reported 19 genes that were significantly mutated in ccRCC (16). It is noteworthy that the enhanced comprehension of the pathologic and signaling anomalies that are distinctive of ADPKD has unveiled distinct parallels with solid tumors.(17). The similarities between ADPKD and cancer are intriguing, particularly given the promising outcomes of recent studies - both preclinical and clinical - that have employed drugs initially designed for treating malignancies (18). ADPKD exhibits several cancer symptoms described by Hanahan and Weinberg in the literature. However, there exist crucial differences between ADPKD and cancer, such as the obligatory preservation of cell polarity, which is a fundamental characteristic of ADPKD, unlike cancer traits such as heightened proliferation or genomic instability. Nonetheless, ADPKD presents some similar defects to those observed in tumors (18). Therefore, the objection of this study is to explore the gene signature shared between ADPKD and RCC based on integrated network analysis.
Methods
The genes associated with ADPKD and renal cancers were downloaded independently. We managed to exclude the overlapped genes. Consequently, the function of common genes was defined and hub genes were detected. The flow chart of this study is shown in Figure 1.

Figure 1. Study workflow. GDA, Gene Disease Association; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DPI, disease probability index.
Data collection
A list of diseases related to autosomal polycystic disease and renal carcinoma was determined. Selected search items were as for ADPKD: “polycystic”, “polycystic kidney”, “autosomal polycystic” and “polycystic kidney disease”; for RCC: “nephron”, “Renal carcinoma”, “renal malignancy”, “clear cell renal carcinoma”, “papillary renal cell carcinoma”, “chromophobe renal cell carcinoma”, which were searched in DisGenNET (19), one of the largest available platform with collected data based on human diseases genetics, (https://www.disgenet.org), then the codes related to that disease were extracted. Moreover, the commonly shared genes were identified via drawing a Venn diagram (20). Of 2797genes related to renal carcinomas of all types and 295 genes related to autosomal dominant polycystic disease, a shared group of 187 genes was identified in common between the two diseases as shown in Figure 2.

Figure 2. Ven diagram of shared genes between ADPKD and renal carcinoma.
To analyze the overlapped genes, we utilized the Enrichr web-based tool (https://maayanlab.cloud/Enrichr) for Gene Ontology assessments in three categories, namely Molecular Function (MF), Biological Process (BP), and Cellular Compartment (CC), and also for identifying Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (21, 22). Significantly enriched Gene Ontology terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, with adjusted P-values lower than 0.05, were identified. Further analysis was conducted on the overlapped genes within the pathways to determine the overrepresented genes within a set of pathways.
To create protein interaction (PPI) networks of the overlapping genes, we utilized the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database (version 11.5) (https://string-db.org), considering a confidence score greater than 0.4. We then employed the cytoHubba plugin (version 0.1) in the Cytoscape software (version 3.8.2) to identify potential hub genes (23, 24).
Results
The selected ADPKD and RCC were searched in DisGeNET and the codes and genes related to them were determined. Following removing duplicates, the pairwise-based analysis identified genetic overlap across the 13 selected disorders (Table 1).
Among 2797 ADPKD-associated genes, 187 genes were shared between ADPKD and RCC. When the genes with DisGeNET score > 0.1 were filtered, 187 genes were detected as common genes among the selected disorders. Remarkably, TGFB1, SNIA2, STAT6, CSF1, and IL6 were the common genes with the highest DisGeNET score.
Table 1. Shared genes of renal cell carcinoma with ADPKD
|
Renal Cell Carcinoma |
Shared genes with ADPKD, n (%) |
|
Clear cell type |
98(52%) |
|
Papillary |
39(20%) |
|
Chromophobe |
50(26%) |
GO and KEGG enrichment analysis
To prospect the potential biological function of 2797 common genes, GO and KEGG pathways enrichment analyses were conducted using the Enrichr, and the component with adjusted P-value ≤ 0.01 were selected. The genes were mainly enriched in the regulation of cell population pathway (GO:0042127) (BP), cytokine-mediated signaling pathway (GO:0019221) (BP), and receptor-ligand activity (GO:0048018) (MF). The 10 top enriched GO terms of overlapped genes between MS and UDs are shown in Figures 3-5. The GO results were consistent with those of the KEGG pathway analysis. For instance, the results revealed that overlapping genes were related to pathways in cancer, the AGE-RAGE signaling pathway in diabetic complications, the PI3k -Akt signaling pathway, and microRNAs in malignancies (Figures 3-5).
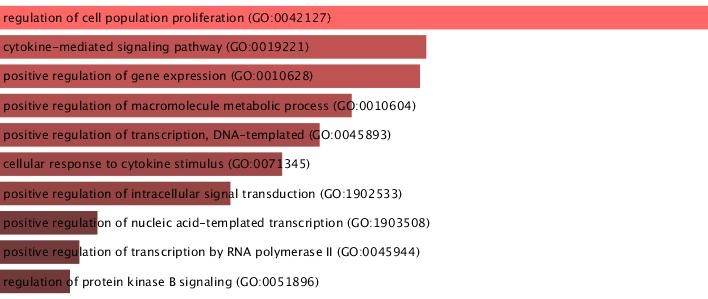
Figure 3. GO biological pathway

Figure 4. GO molecular function

Figure 5. GO cellular function
To prioritize candidate common genes, we further categorized them by the rank of disease-associated pathways that they are involved. Then, we ranked the 20 top genes for each GO term and pathway and performed a pairwise analysis (Table 1). According to our analysis, TNF, IL6, TGFB1, and, TGFA, EPO overlapped among biological process and molecular function terms, and KEGG pathway.
Table 2. Go enrichment analysis of overlapped genes between ADPKD and RCC
|
GO_Biological_Process |
Go_Molecular_Function |
GO_Cellular_Component |
KEGG-Pathway |
|
CXCL9 |
TNF |
SPARC |
PIK3CB |
|
CSF1 |
CSF1 |
CSF1 |
PIK3CD |
|
EPO |
EPO |
COL14A1 |
EPO |
|
MYC |
CXCL9 |
HP |
EDNRA |
|
TGFB1 |
TGFB1 |
TGFB1 |
TGFB1 |
|
TIMP1 |
EDN1 |
ETFA |
TNF |
|
JUNB |
PTH |
CSF1 |
IL6 |
|
JAK2 |
EGF |
JAK2 |
JAK2 |
|
EDN1 |
IL18 |
EGFR |
EDN1 |
|
TGFA |
TGFA |
GLS |
TGFA |
|
ADAM10 |
MIF |
CST3 |
TP53 |
|
TSC2 |
TNF |
MUC1 |
EGFR |
|
TSC1 |
AGT |
ERBB4 |
ERBB2 |
|
RUNX1 |
VEGFA |
MAPK1 |
MAPK1 |
|
SFRP4 |
LGALS3 |
APOE |
EDNRB |
|
IL6 |
GH1 |
IL6 |
TGFB2 |
|
AGTR1 |
CXCL10 |
PTGS2 |
STAT5B |
|
TP53 |
IL6 |
MIF |
SMAD2 |
|
TNF |
SST |
FGF23 |
HMOX1 |
|
AVPR2 |
IL1B |
TNF |
IL13 |
Identification of hub genes via PPI network
The PPI network of overlapped genes was constructed with 985 nodes using STRING. The degree was calculated using the cytoHubba plugin in Cytoscape software and the top 20 ranking genes, including AXIN1, MYCN, TWIST1, MMP11, AGTR, CASP1, STAT6, TNF, IL6, CSF, SNAI1, TGFB1, JAK2 were identified as hub genes (Figure 6).

Figure 6. PPI network and common hub genes
The hub genes were also searched among the genes which were determined as key genes with the highest over presentations in disease-associated pathways (Figure 7). According to our analysis, TNF, TGFB1 and JAK2, EPO, CSF1, and TGFA were detected as the overrepresented genes in biological process and molecular function terms as well as KEGG.

Figure 7. The hub genes among the genes, which were determined as keygens with the highest over presentations in disease-associated pathways
Discussion
We identified five hub genes between ADPKD and RCC based on integrated network analysis including TGFB1, SNIA2, STAT6, CSF1, and IL6. Numerous studies have confirmed that a wide variety of pathological pathways are involved in ADPKD. An integrative bioinformatics study on molecular pathway of CKD by Zhou LT, and colleagues indicated to numerous key genes (e.g. OAS1, JUN, and FOS) can play critical roles in the progression of CKD (25). Transcriptomic profiles of different types of CKD indicated several previously known key CKD genes (e.g., TGFBI, COL4A1, and FCN1 (25). TGF-beta (transforming growth factor-beta) is one of our five-candidate genes between ADPKD and RCC. Systematically, TGF-β signaling, together with histone deacetylase 7 (HDAC7), suppresses tricarboxylic acid cycle, (TCA cycle), also called Krebs cycle expression. (26).
The PPI network encoded by the common DEGs showed ten hub proteins (EPHB2, PRKAR2B, CAV1, ARHGEF12, HSP90B1, ITGA2B, BCL2L1, E2F1, TUBB1, and C3) identified common gene signature and pathways in COVID-19 patients with CKD comorbidity (27). A bioinformatics analysis and machine learning on CKD and non-alcoholic fatty liver disease (NAFLD) suggested four NAFLD-related genes (DUSP1, NR4A1, FOSB, ZFP36) as diagnostic markers in CKD patients with NAFLD. Several studies indicated different genes in IgA nephropathy (IgAN) (28, 29). IL10, IGLL5 and POU2AF1 genes were selected as key genes and their targets were recognized as MAPK1 and PPKCA in the biomarkers associated with RCC microenvironment with therapeutic and prognostic value. A positive correlation between IL/POU2AF1 expression and abundance of six immune cells was observed (30).
The study by Grimaldi AM., as a comparative bioinformatic analysis on two major subtypes of RCC showed the importance of the CBX gene family. CBX1, CBX6, and CBX7 were also expressively related to the tumor stage. Additionally, decreased expression levels of CBX1, CBX5, CBX6, CBX7, and increased expression of CBX8 were associated with poor prognosis (31). High mRNA expression of cyclin D1 (CCND1), fms-related tyrosine kinase 1 (FLT1), plasminogen (PLG), and von Willebrand factor (VWF) are important in recurrent free survival of RCC patients (32). Also, Using bioinformatics analysis exposed that Fibroblast Growth Factor 1 (FGF1) expression was abnormally lost in ccRCC which statistically suggestively connected to the patient's overall survival (OS) (33).
Conclusion
In 187 common genes between ADPKD and RCC, the final 5 key genes TGFB1, SNIA2, STAT6, CSF1, and IL6 were mutual across. The identified feature could be potential targets for ADPKD patients at risk of RCC.
Acknowledgments
Special thanks to Sina Hospital, Tehran University of Medical Sciences, Tehran, Iran.
Competing interests
The authors declare no Conflict of Interest for this article.
Founding
There is no funding
Ethical statement
Not Applicable
Data Availability
All necessary data are included in the manuscript and no additional data are included.
Abbreviations
ADPKD Autosomal dominant polycystic kidney disease
BP Biological process
CC Cellular compartment
CKD Chronic Kidney Disease
CNAs Copy number alterations
MF Molecular function
NAFLD Non-alcoholic fatty liver disease
TME Tumor microenvironmental
VHL Von Hippel-Lindau
VWF Von Willebrand factor